A multidisciplinary team of roboticists, ocean engineers, bioengineers, and marine and molecular biologists has successfully demonstrated how new technologies can rapidly obtain high-resolution 3D images and preserved tissue to accelerate the discovery of new life in the deep sea.
The research team—led by the University of Rhode Island (URI) in collaboration with MBARI, Bigelow Laboratory for Ocean Sciences, Harvard University, PA Consulting, and Baruch College—published their findings in Science Advances today. The team has shown that it is possible to shave years from the process of determining whether a new or rare species has been discovered. Their findings represent five years of work to find faster ways to collect detailed information about deep-sea animals.

MBARI Principal Engineer Kakani Katija (right) joined a team of roboticists, engineers, and biologists led by the University of Rhode Island to demonstrate how technology can accelerate research on delicate deep-sea animals. (Image credit: Jovelle Tamayo, 2021 Schmidt Ocean Institute)
“The ocean is at a critical crossroads. We urgently need to establish a baseline of who lives in the deep sea so we can assess and track the impacts of human activity across all ocean ecosystems. Advanced imaging technologies can accelerate this work, helping us better understand marine life so we can better protect ocean animals and environments for future generations,” said MBARI Principal Engineer Kakani Katija, a coauthor of the study.
Revolutionary advancements in underwater imaging, robotics, and genomic sequencing are transforming ocean exploration. By leveraging these new tools, within minutes of an encounter with a deep-sea animal, scientists can capture detailed measurements and motion of the animal, obtain its entire genome, and generate a comprehensive list of genes being expressed to assess its physiological status. The research team calls the result of these rich digital data a “cybertype” of a single animal rather than a physical holotype specimen that is traditionally found in museum collections.
When researchers encounter what they believe to be a new species, they first face the challenge of collecting an intact specimen and transporting it to the surface. Describing a new species requires detailed analysis of defining physical features, requiring the specimen to be preserved for further study. Some organisms, especially gelatinous animals, are so delicate that their tissues cannot withstand preservation agents. The cybertype can fill in the gaps for specimens that break down over time, ensuring that information about these species is available for future scientists.
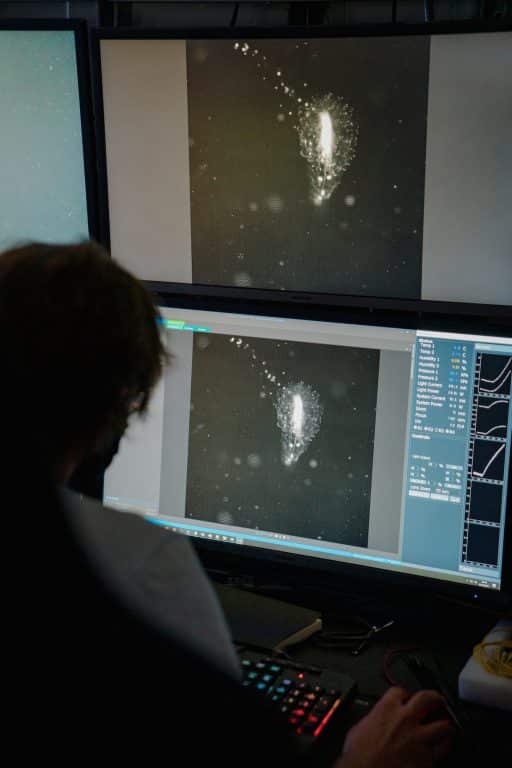
Imaging systems developed by MBARI’s Bioinspiration Lab can help biologists visualize the delicate tissues of gelatinous animals. (Image credit: Jovelle Tamayo, 2021 Schmidt Ocean Institute)
“Currently, if researchers want to describe what they believe is a new species, they face an arduous process,” said URI Professor Brennan Phillips, the principal investigator on the project. Phillips noted that the backlog of describing new species has hampered conservation efforts. “When you look at climate change and deep-sea mining and their potential effects, it is unsettling. You realize you don’t have a full baseline of species, and you may not know what you’ve lost before it’s too late. If you want to know what has been there before it’s gone, this is a new way to do that.”
Two expeditions with the Schmidt Ocean Institute aboard the research vessel Falkor offshore of Hawaii in 2019 and off the coast of San Diego in 2021 allowed Phillips, Katija, and team to test technology for creating a cybertype for deep-sea animals.
The team collected as many as 14 preserved tissue samples a day, along with terabytes of quantitative digital imagery. Together, the study provided:
- The first complete assembled and annotated transcriptome (genes being made in the animals’ habitat) of the salp Pegea, a barrel-shaped gelatinous invertebrate;
- Details of the molecular basis of environmental sensing of the gossamer worm (Tomopteris sp.), a bristle worm that lives in the water column;
- Details of the full transcriptomes of two gelatinous siphonophores, Erenna sp. and Marrus claudanielis, as well as Pegea and Tomopteris;
- Full morphological (form and structure) characterizations using digital imaging of each animal while at depth.
Advanced imaging technology developed by MBARI’s Bioinspiration Lab was crucial to this effort.
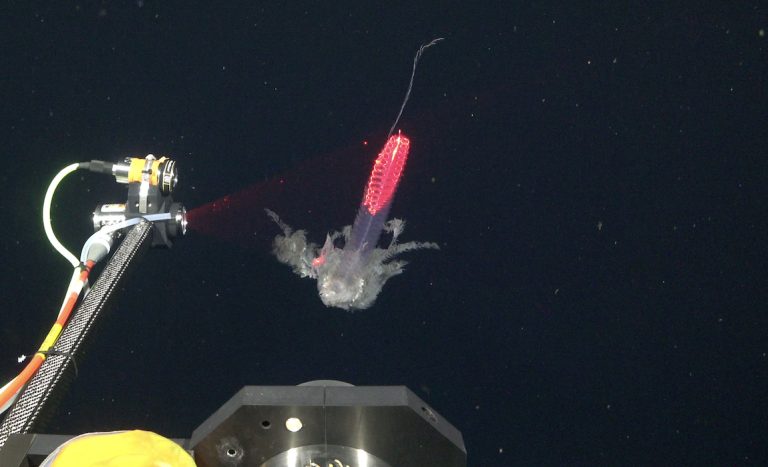
MBARI’s DeepPIV imaging system projects a sheet of laser light to take a detailed scan of delicate deep-sea animals so researchers can build digital recreations of their fine structures. (Image credit: Schmidt Ocean Institute)
Katija and her team in the Bioinspiration Lab work to bring the lab to the ocean, creating new high-tech tools to study how deep-sea animals survive in a deep, cold, and dark environment. Ultimately, the team hopes to help find bioinspired solutions to some of the world’s biggest engineering challenges.
The Bioinspiration Lab has developed innovative imaging systems that can visualize deep-sea animals in their natural environment. Non-invasive scans with lights and lasers gather data that can be used to create a three-dimensional model of an organism.
Mounted on a remotely operated vehicle (ROV), the Deep Particle Imaging Velocimetry (DeepPIV) instrument projects a sheet of laser light that illuminates particles in the water, like dust in a sunbeam. The laser scan creates 3D in situ visualizations of the structures of soft-bodied deep-sea organisms. By recording the movement of particles surrounding an organism, researchers can also quantify the tiny currents around marine animals created as they eat or move. MBARI researchers have previously used this technology to visualize the “snot palaces” of giant larvaceans, tadpole-like animals crucial in cycling carbon from the surface to the seafloor.
The three-dimensional lightfield (plenoptic) EyeRIS camera system can image particle and tissue movement. Funded by the Gordon and Betty Moore Foundation and developed by MBARI’s Bioinspiration Lab, this instrument allows researchers to quantitatively study the form and function of organisms, small-scale fluid dynamics, and particle fields, all in three dimensions.

The EyeRIS camera system developed by MBARI engineers captures detailed information about an animal’s physical appearance and movement in three dimensions. (Image credit: Schmidt Ocean Institute)
“New imaging technologies developed at MBARI are providing an unprecedented view of ocean life. By outfitting underwater robots with advanced camera systems, we can now observe animals in their natural environment and learn about their complex body structures and behaviors without bringing them to the lab,” explained Katija.
After collecting in situ morphological data using the ROV’s ultra high-definition 4K science camera and the EyeRIS and/or DeepPIV imaging systems, Phillips and his collaborators leveraged a groundbreaking sampling system to collect genomic information about deep-sea animals in their natural habitat hundreds of meters below the surface.
Developed by engineers in the School of Engineering and Applied Sciences at Harvard University, the rotary-actuated dodecahedron (RAD-2) folds around an animal like origami to collect the specimen and sample its tissues. This sampler preserves a snapshot of the organism’s genetic activity at the time of collection, allowing scientists to identify which genes were active and being expressed at that time, known to scientists as the transcriptome.

The rotary-actuated dodecahedron (RAD-2) developed by Harvard University engineers folds around an animal like origami to collect the specimen and sample its tissues. (Image credit: Schmidt Ocean Institute)
The team envisions using these advanced technologies to build a comprehensive picture of the biology and behavior of deep-sea animals. For example, they observed a gossamer worm (Tomopteris sp.)—a worm that lives in the water column instead of along the seafloor—while it was swimming back and forth exploring its surroundings. Rapid collection and preservation of the worm’s tissues revealed high levels of gene expression in the whisker-like cirri, suggesting these organs serve a neurosensory function for following chemical trails.
Further development of these technologies could eventually allow researchers to collect complete scans and inventories of life in the deep sea within a catch-and-release framework.
“By combining a suite of emerging technologies, we’re transforming our ability to explore the ocean. Eventually being able to conduct quantitative observations of ocean life while being completely non-invasive will be a game changer moving forward. Not only will we be able to study species that have historically been too difficult to collect, but all of that digital data will be easily shareable and accessible. The ocean is changing. We need to expand access to ocean data because we need all hands on deck to protect marine life and ecosystems at this critical time,” said Katija.
This work was funded by the Schmidt Ocean Institute, the David and Lucile Packard Foundation, the Gordon and Betty Moore Foundation (grant #7583), the National Science Foundation (NSF OIA-1826734), and the National Geographic Society (grant no. SP 12-14).

